Abstract
Introduction
Graft-versus-host disease (GvHD) is the major cause of non-relapse mortality in allogeneic hematopoietic cell transplantation (alloHCT), and about 40-60% cases would progress into steroid refractory (SR) aGvHD resulting in dismal outcomes. The pathophysiology of aGvHD is complex, and donor derived T cells are identified as the culprits of aGvHD. However, the relationships among distinct T cell subsets and molecular mechanisms underlying aGvHD remain incompletely understood.
Methods
We employed a single-cell RNA sequencing (scRNA-seq) and T cell receptor (TCR) sequencing (scTCR-seq) to profile 49288 T cells in peripheral blood isolated from alloHCT recipients developed into aGvHD (n=9) or not (as control, n=5). Then, samples from SR-aGvHD patients were further investigated to explore the molecular mechanism of pathogenic T cell subsets in SR-aGvHD.
Results
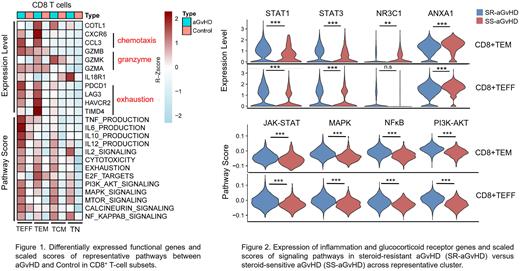
A total of thirteen unique T cell subsets with distinct molecular properties were identified, including four clusters of CD8+ T cells (CD8+ TN, CD8+ TCM, CD8+ TEM and CD8+ TEFF), five clusters of CD4+ T cells (CD4+ TN, CD4+ TCM, Th1, Th17 and Treg), NK cell cluster and three clusters of other T cells (IFN-T, γδT and MAIT). The composition shifted towards more-developed CD8+ TCM, CD8+ TEM, CD4+ TCM and Th1 cells in aGvHD group due to rapid cell cycling. The expansion of CD8+ TEM subpopulation was confirmed by multiparameter flow cytometry. The CD8+ TEFF and Th1 were considered to be pathogenic in aGvHD as they exhibited aGvHD-related cytokine/granzyme expression and signaling pathway activation. Upregulated genes in aGvHD-derived-Th1 cells were related to chemotaxis (CCL3, CXCR6), cytokine production (type I interferon, IL-23) and JAK-STAT signaling activation. CD8+ TEFF cells showed activity of chemotaxis (CCL3, CXCR6), granzyme and cytokine expression (GZMB, GZMA, TNF-α, IL-6, IL-10, IL-12) and MAPK/MTOR/NF-kappaB activation. The different composition of cytokine expression and activated signaling pathways suggested distinct functions of each subset. Furthermore, CD8+ TEM represented four traits related to a range of T-cell differentiation stages, spanning rapid cell cycling (transcription factor E2F family), chemotaxis (CCL3, CXCR6), cytotoxicity (GZMA, GZMB, and GZMK) and exhaustion (PDCD1, LAG3, HAVCR2 and TIMD4), suggesting CD8+ TEM as a transient stage which might further develop into CD8+ TEFF for aGvHD pathogenesis (Figure 1).
To elucidate the molecular mechanism of SR-aGvHD, we dissected the composition and functional heterogeneity of T-cell subsets between SR- and steroid sensitive (SS)-aGvHD. CD8+ TEM expanded to be the largest subset which resulted in the increase of CD8+ TEFF proportion in SR-aGvHD. CD8+ TEM and CD8+ TEFF showed enrichment of GvHD pathogenic scores and up-regulated expression of inflammation transcription factors STAT1 and STAT3 in SR-aGvHD. Of note, the expression of steroid receptor NR3C1 was downregulated in CD8+ TEM subset, as well as the steroid target gene ANXA1, compared with counterparts. Furthermore, both CD8+ TEM and CD8+ TEFF cells exhibited enrichment of JAK-STAT, MAPK, NF-kappaB, and PI3K-AKT signaling activation (Figure 2).
Conclusions
The effector T-cell subsets CD8+ TEFF and Th1 were co-activated and undertook distinct expression profiles and signaling pathways, which suggests their different roles in aGvHD pathogenesis. CD8+ TEM, characterized by proliferative, cytotoxicity, chemotaxis and exhaustion capacities, provided functional repertoire for CD8+ TEFF. Reduced NR3C1 expression and activation of by-pass survival pathways of CD8+ TEM might provide persistent differentiation origin of CD8+ TEFF to mediate resistance to steroid therapy. Collectively, our data described the immune landscape of aGvHD and provided novel therapeutic targets.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal